Michigan Medicine - University of Michigan
Summary
Adipose tissue macrophage (ATM) infiltration is associated with adipose tissue dysfunction and insulin resistance in mice and humans. Recent single-cell data highlight increased ATM heterogeneity in obesity, but do not provide spatial context for the dynamics of the ATM phenotype. We integrated single-cell RNA-Seq, spatial transcriptomics, and murine adipose tissue imaging in a temporal study of diet-induced obesity. Overall, pro-inflammatory immune cells predominated in early obesity, while non-resident anti-inflammatory ATMs predominated in chronic obesity. A subset of these anti-inflammatory ATMs were transcriptomically intermediate between monocytes and lipid-associated mature macrophages (LAM) and were consistent with a LAM precursor (pre-LAM). Pre-LAMs were spatially associated with crown-shaped structures (CLS) of early obesity, indicating adipose tissue dysfunction. Spatial data showed colocalization of ligand-receptor transcripts related to lipid signaling between monocytes, pre-LAM and LAM, including Apoe, Lrp1, Lpl and App. Pre-LAM expression of these ligands in early obesity suggested signaling to LAM in the CLS microenvironment. Our results refine the understanding of adipose tissue macrophage (ATM) diversity and provide insights into LAM lineage dynamics during metabolic disease development. |
Comments
Fat tissue , as much as it has been vilified, is an incredibly complex and essential body organ involved in energy storage and hormone production, among other functions. However, modern lifestyles have led to a global epidemic of obesity and a corresponding rise in related conditions such as type 2 diabetes and cardiovascular disease.
Researchers are trying to uncover the basics of how adipose tissue is structured, and specifically the inflammation associated with obesity , in hopes of uncovering the connection between fat accumulation and poor health outcomes.
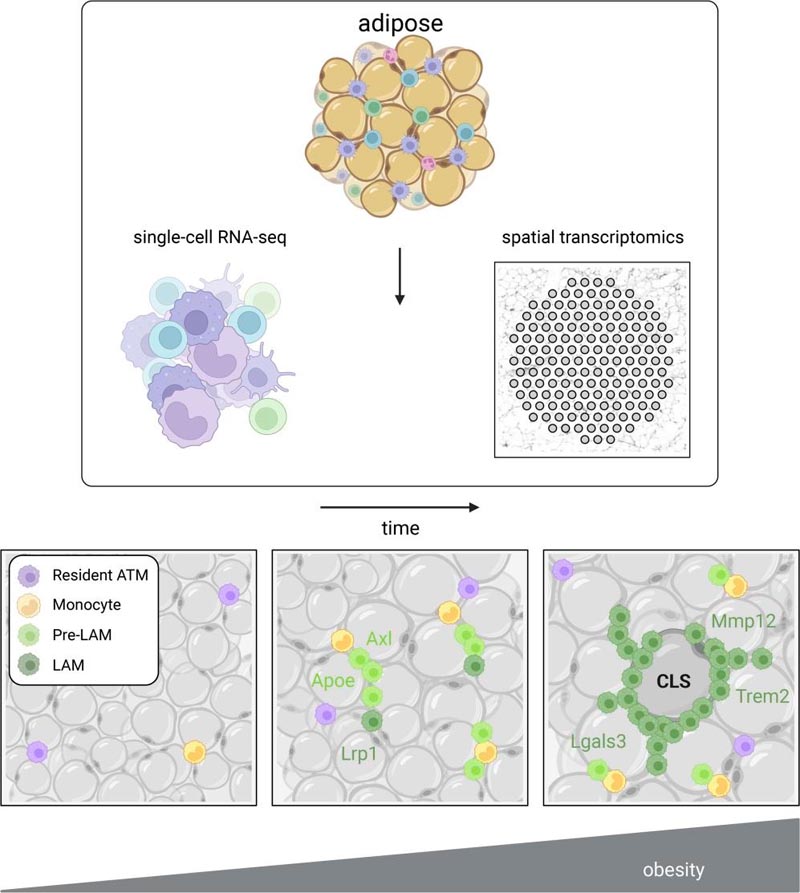
A new study by Lindsey Muir, Ph.D., doctoral candidate Cooper Stansbury, and colleagues uses single-cell analysis of gene expression combined with spatial transcriptomics to reveal previously unrecognized immune cell types and interactions within adipose tissue. Spatial transcriptomics is a newer technology that captures all gene expression in small spots in an entire thin section of tissue.
Studying fat is easier said than done. In tissues that are organized in defined layers, for example the spinal cord or the brain, “it is easier to do checks with your data and identify this or that layer as a particular cell type and know that it should express genes X, Y and Z,” said Muir, a research assistant professor in the Department of Computational Medicine and Bioinformatics.
"With adipose tissue, it’s much more difficult because the cell types are evenly distributed throughout the tissue, without defined cell layers." In obesity, fat cells or adipocytes expand and can reach a limit which ultimately causes cell death and leads to inflammation.
To better understand the types of immune cells within adipose tissue and where they meet each other in obesity, the team fed mice a high-fat diet for 14 weeks, harvested fat tissue, and then used spatial and single-cell analyzes to produce a readout of all the mRNAs present in the sample.
Using a computational process known as clustering on single-cell data , they were able to group together cells whose genetic makeup was more similar to each other than other groups or the overall sample.
They discovered something surprising about the samples’ macrophage population , an immune cell whose job is to clean up dead cells and debris.
"We knew that macrophages would probably have multiple subtypes, what surprised us was the number that emerged, which were very different from each other and appeared at different times and became more dominant over time."
They identified five types, which they called Mac1, 2, 3, 4 and 5. Mac1 resided in the tissue of both lean mice on a normal diet and obese mice. Mac2 and Mac3, which were identified by their pro-inflammatory genes , peaked after 8 weeks of high-fat diet.
However, as the high-fat diet progressed to 14 weeks, Mac4 and Mac5 cells, which had low expression of pro-inflammatory genes, predominated, while pro-inflammatory Mac2 and Mac3 cells decreased.
“The idea in the field has been that the type of macrophages that accumulate in obesity promote an inflammatory state . Based on this data, there is a lot more to the story,” Muir said.
Their hypothesis is that Mac4 and Mac5 are the lipid-associated macrophages (LAM) described in their own previous work and by other researchers and may be a signal that the body is trying to quell a harmful level of inflammation coming from pro-inflammatory macrophages and dying adipocytes. .
Next, painstakingly careful sectioning of fresh frozen fat tissue enabled analysis by spatial transcriptomics. Each analysis point in the spatial method has a unique barcode that is attached to the mRNA in the tissue above that point, so gene expression can subsequently be mapped to specific locations in the tissue using the barcodes as coordinates. In this method, sections are also imaged just before mRNA collection. The study examined these images for telltale markers called crown-shaped structures , structures that are associated with insulin resistance .
"Once crown -like structures appear , they take a long time to disappear and their appearance indicates tissue dysfunction," Muir said. "Using image processing, we identified, based on the density of these regions, what would likely be a crown-like structure and then verified that we could see them visually," Muir said. These structures had gene expression indicating the presence of LAM Mac4 and Mac5.
With more information about the cellular composition and spatial organization of adipose tissue in the context of obesity, the next step, Muir said, is to examine the signaling processes and proteins associated with the development of LAM and metabolic disorders.
Conclusions Our data review the current understanding of ATM phenotypic changes in obesity. We identify important milestones in the development of MN-LAM and provide a spatial context for myeloid signaling implicated in metabolic dysfunction . Our study provides clarity on the cell types and signaling involved in the formation and accumulation of crown-shaped structures (CLS), including the spatial dynamics of LAM development in obesity. |
Reference : “ A lipid-associated macrophage lineage rewires the spatial landscape of adipose tissue in early obesity ”, JCI Insight, DOI: /10.1172/jci.insight.171701