Structured summary
Neuropsychiatric disorders and mental illnesses are the leading cause of disease burden in the United States. Tens of thousands of sequence variants in the human genome have been linked to the etiology of these conditions. However, elucidating the role of the identified risk variants remains challenging because most of them are located outside protein coding regions and currently lack functional annotation. These disease risk variants are likely to exert their influence by perturbing transcriptional regulatory elements, thereby modulating gene expression in cell types relevant to neuropsychiatric disorders. Recent advances in single-cell technologies have revealed a high degree of cellular heterogeneity throughout the human brain. However, the transcriptional regulatory sequences that govern the identity and function of each individual brain cell type have not yet been delineated, hampering our ability to interpret non-coding disease risk variants.
Fundamental reason
Conventionally, transcription regulatory sequences can be determined by evidence of chromatin accessibility that typically accompanies transcription factor binding and chromatin remodeling. However, previous catalogs of transcriptional regulatory elements lack information on the cell type-specific activities of each element due to the use of bulk tissue samples. Recent technological advances have allowed us to analyze chromatin accessibility at the single-cell level, allowing the creation of cell type-specific maps of transcriptional regulatory elements for complex organs such as the human brain.
Results
In this study, we present a comprehensive analysis of chromatin accessibility in the human brain at the single-cell level, encompassing a collection of 1.1 million cells from 42 distinct brain regions in three neurotypical adult subjects. We used this chromatin atlas to define 107 distinct brain cell types and discovered the chromatin accessibility status of 544,735 putative transcriptional regulatory elements in these cell types.
A substantial number of these regulatory elements also exhibited sequence conservation and chromatin accessibility in mouse brain cells , underscoring their functional importance. Through integrative analysis, we have linked many putative transcriptional regulatory elements to potential target genes. Additionally, we leveraged this atlas to predict cell types relevant to 19 neuropsychiatric traits and disorders. Finally, we developed machine learning models to predict the regulatory function of disease risk variants. We have made this atlas available to the public free of charge through the interactive web portal CATLAS (www.catlas.org).
Conclusions The single-cell chromatin atlas of the human brain represents a valuable resource for the neuroscience community. It provides insights into the gene regulatory programs that shape the diversity of brain cell types and helps interpret the functional roles of disease risk variants located outside protein-coding regions. This atlas, in combination with other molecular and anatomical data, promises to advance our understanding of brain function and neuropathology and ultimately offers avenues for more effective approaches to address neuropsychiatric disorders . |
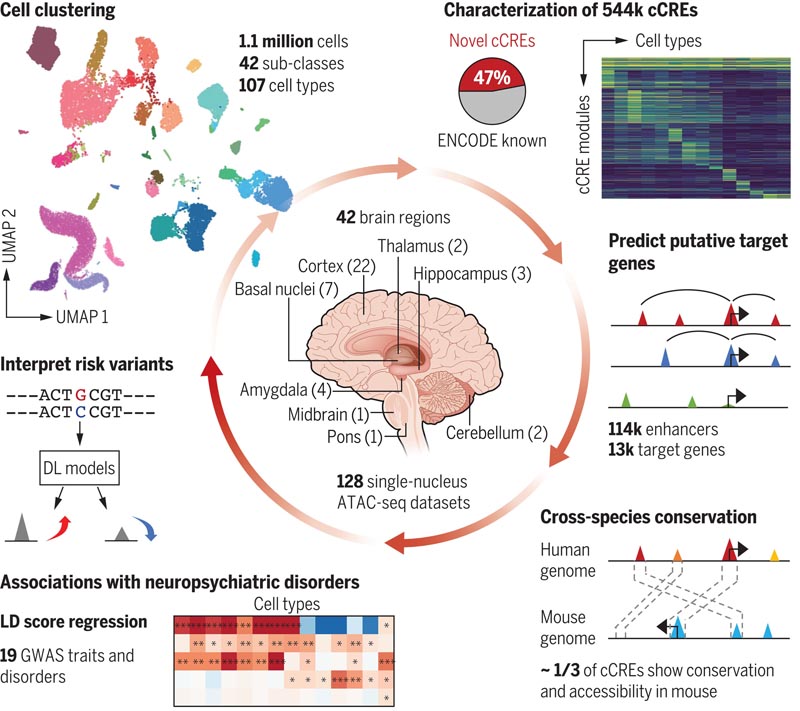
Figure: Single-cell analysis of human brain chromatin accessibility . Candidate cis-regulatory elements (cCREs) specific to different human brain cell types were identified using a single-core assay for transposase-accessible chromatin by sequencing (snATAC-seq) and linked to putative target genes by integrative analysis. The use of cCRE was leveraged to predict brain cell types relevant to neuropsychiatric traits and disorders and to train machine learning models to interpret the function of non-coding risk variants. UMAP, Multiple Uniform Approach and Projection; DL, deep learning; LD, linkage disequilibrium; GWAS, genome-wide association study.
Comments
Researchers map genetic switches and brain cell types associated with schizophrenia, bipolar disorder, Alzheimer’s disease and major depression.
In a large multi-institutional effort led by the University of California, San Diego, researchers analyzed more than one million human brain cells to produce detailed maps of genetic switches in brain cell types and revealed links between specific cell types and several factors. common to neuropsychiatric disorders. The team also developed artificial intelligence tools to predict the influence of individual high-risk genetic variants among these cells and how they may contribute to disease.
The new work, published in a special issue of Science , is part of the National Institutes of Health’s Brain Research through Advancing Innovative Neurotechnologies Initiative, or BRAIN Initiative, launched in 2014. The initiative aims to revolutionize understanding of the mammalian brain, in part, by developing new neurotechnologies to characterize neuronal cell types.
Each human brain cell contains the same DNA sequence, but different types of cells use different genes and in different quantities. This variation produces many different types of brain cells and contributes to the complexity of neural circuits. Learning how these cell types differ at a molecular level is critical to understanding how the brain works and developing new ways to treat neuropsychiatric diseases.
"The human brain is not homogeneous ," said senior author Bing Ren, PhD, a professor at UC San Diego School of Medicine. "It is made up of a hugely complex network of neurons and non-neuronal cells, each of which fulfills different functions. Mapping the different types of cells in the brain and understanding how they work together will ultimately help us discover new therapies that can target to individuals cell types relevant to specific diseases.
In the new study, researchers analyzed more than 1.1 million brain cells in 42 different brain regions of three human brains. They identified 107 different subtypes of brain cells and were able to correlate aspects of their molecular biology with a wide range of neuropsychiatric diseases, including schizophrenia, bipolar disorder, Alzheimer’s disease and major depression . Researchers then use this data to create machine learning models to predict how certain sequence variations in DNA may influence gene regulation and contribute to disease.
While these new results offer important information about the human brain and its pathology, scientists are still far from finished with brain mapping. In 2022, UC San Diego joined the Salk Institute and others to launch a Multi-Omic Atlas Center of Human Brain Cells, which aims to study cells from more than a dozen human brains and raise questions about how the brain changes during development. throughout people’s lives and with illness.
"Expanding our work to an even greater level of detail in a larger number of brains will bring us one step closer to understanding the biology of neuropsychiatric disorders and how they can be rehabilitated," Ren said.
Lead authors of the study include: Yang Eric Li, Sebastian Preissl, Michael Miller, Zihan Wang, Henry Jiao, Chenxu Zhu, Zhaoning Wang, Yang Xie, Olivier Poirion, Colin Kern, Lin Lin, Qian Yang, Quan Zhu, Nathan Zemke. , Sarah Espinoza, Jingbo Shang and Allen Wang at UC San Diego, Nicholas D. Johnson Antonio Pinto-Duarte, Wei Tian Nora Emerson, Julia Osteen, Jacinta Lucero, M. Margarita Behrens and Joseph R. Ecker at the Salk Institute for Biological Studies , Kimberly Silett and Sten Linnarsson of the Karolinksa Institute Anna Marie Yanny, Julie Nyhus, Nick Dee, Tamara Casper, Nadiya Shapovalova, Daniel Hirschstein, Rebecca D. Hodge Trygve Bakken, Boaz Levi and Ed Lein of the Allen Institute for Brain Sciences and C. Dirk Keene at the University of Washington Seattle.
The study was supported by the National Institutes of Health (grants UM1MH130994, U01MH114812, U54HG012510, and S10 OD026929), the National Science Foundation (grant OIA-2040727); the Nancy and Buster Alford Endowment, the Life Sciences Research Foundation, as well as donations from Google, Adobe and Teradata.